Among the myriad of problems we are having with the COVID-19 pandemic, faster testing is one we could actually improve. The standard test for the presence of SARS-CoV-2 virus uses PCR (polymerase chain reaction), which amplifies targeted viral RNA. It is accurate (high specificity) but requires relatively expensive equipment and reagents that are currently in short supply. There are reports of wait times of over a week, which renders a test useless for contact tracing.
An alternative to PCR is an antigen test that tests for the presence of protein fragments associated with COVID-19. These tests can in principle be very cheap and fast, and could even be administered on paper strips. They are generally much more unreliable than PCR and thus have not been widely adopted. However, as I show below by applying the test multiple times, the noise can be suppressed and a poor test can be made arbitrarily good.
The performance of binary tests are usually gauged by two quantities – sensitivity and specificity. Sensitivity is the probability that you test positive (i.e are infected) given that you actually are positive (true positive rate). Specificity is the probability that you test negative if you actually are negative (true negative rate). For a pandemic, sensitivity is more important than specificity because missing someone who is infected means you could put lots of people at risk while a false positive just means the person falsely testing positive is inconvenienced (provided they cooperatively self-isolate). Current PCR tests have very high specificity but relatively low sensitivity (as low as 0.7) and since we don’t have enough capability to retest, a lot of tested infected people could be escaping detection.
The way to make any test have arbitrarily high sensitivity and specificity is to apply it multiple times and take some sort of average. However, you want to do this with the fewest number of applications. Suppose we administer 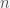 tests on the same subject, the probability of getting more than
tests on the same subject, the probability of getting more than  positive tests if the person is positive is
positive tests if the person is positive is  , where
, where  is the cumulative distribution function of the Binomial distribution (i.e. probability that the number of Binomial distributed events is less than or equal to
is the cumulative distribution function of the Binomial distribution (i.e. probability that the number of Binomial distributed events is less than or equal to  ). If the person is negative then the probability of
). If the person is negative then the probability of  or fewer positives is
or fewer positives is  . We thus want to find the minimal
. We thus want to find the minimal 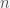 given a desired sensitivity and specificity,
given a desired sensitivity and specificity,  and
and 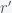 . This means that we need to solve the constrained optimization problem: find the minimal
. This means that we need to solve the constrained optimization problem: find the minimal 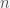 under the constraint that
under the constraint that 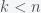 ,
,  and
and 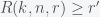 .
. 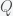 decreases and
decreases and 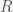 increases with increasing
increases with increasing  and vice versa for
and vice versa for 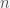 . We can easily solve this problem by sequentially increasing
. We can easily solve this problem by sequentially increasing 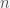 and scanning through
and scanning through  until the two constraints are met. I’ve included the Julia code to do this below. For example, starting with a test with sensitivity .7 and specificity 1 (like a PCR test), you can create a new test with greater than .95 sensitivity and specificity, by administering the test 3 times and looking for a single positive test. However, if the specificity drops to .7 then you would need to find more than 8 positives out of 17 applications to be 95% sure you have COVID-19.
until the two constraints are met. I’ve included the Julia code to do this below. For example, starting with a test with sensitivity .7 and specificity 1 (like a PCR test), you can create a new test with greater than .95 sensitivity and specificity, by administering the test 3 times and looking for a single positive test. However, if the specificity drops to .7 then you would need to find more than 8 positives out of 17 applications to be 95% sure you have COVID-19.
using Distributions
function Q(k,n,q)
d = Binomial(n,q)
return 1 – cdf(d,k)
end
function R(k,n,r)
d = Binomial(n,1-r)
return cdf(d,k)
end
function optimizetest(q,r,qp=.95,rp=.95)
nout = 0
kout = 0
for n in 1:100
for k in 0:n-1
println(R(k,n,r),” “,Q(k,n,q))
if R(k,n,r) >= rp && Q(k,n,q) >= qp
kout=k
nout=n
break
end
end
if nout > 0
break
end
end
return nout, kout
end